Classification with B-Spline Basis Function Coefficients¶
Note
Last updated 11 AM, 9/7/2020
import pickle as pkl
import numpy as np
import pandas as pd
import torch
from torch import nn, optim
from torch.utils.data import Dataset, TensorDataset, DataLoader
import torch.nn.functional as F
from torch.nn.utils.rnn import pack_padded_sequence, pad_packed_sequence
from sklearn.model_selection import KFold
from sklearn.metrics import confusion_matrix
import plotly.graph_objects as go
from plotly.subplots import make_subplots
from plotly.express.colors import cyclical
import plotly.io as pio
pio.renderers.default = 'notebook'
from IPython.display import display, clear_output
from ipythonblocks import BlockGrid
from webcolors import name_to_rgb
from scipy import interpolate
from sympy import lambdify, bspline_basis_set, symbols
from sympy.abc import t
import warnings
warnings.filterwarnings('ignore')
grid = BlockGrid(15,1,fill=(0,0,0))
grid.block_size = 50
grid.lines_on = False
colors = ['slategray','sienna','darkred','crimson','darkorange','darkgoldenrod','darkkhaki','mediumseagreen','darkgreen','darkcyan','cornflowerblue','mediumblue','blueviolet','purple','hotpink']
i = 0
for block in grid:
color = name_to_rgb(colors[i])
block.set_colors(color[0],color[1],color[2])
i+=1
with open('trajectories_updated.pkl', 'rb') as f:
data = pkl.load(f)
traj_df = data['traj_df']
mean_df = data['mean_df']
clipdata_df = data['clipdata_df']
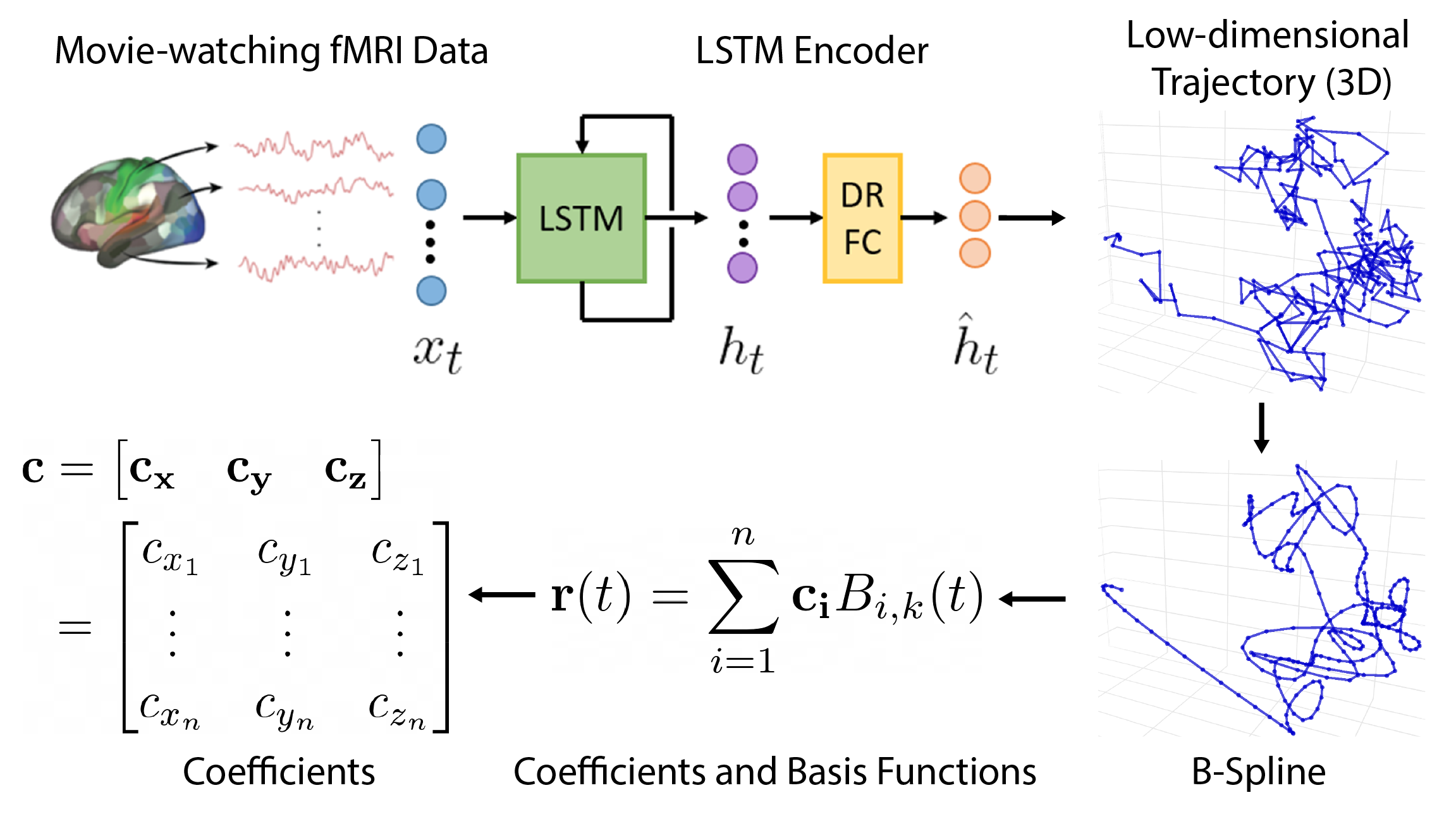
B-Splines¶
From the previous section, we have shown that trajectories can be effectively smoothed with B-splines.
A key concept behind B-splines is that any B-spline can be created from a linear combination of basis functions. Additionally, given the same knot vector and spline order/degree, the B-spline basis functions will always be the same. These two properties mean that the coefficients of the basis functions are sufficient to describe a B-spline.
Here’s a quick overview of how coefficients are calculated.
Bases¶
Let’s take a look at some trajectories and their bases. We’ll use a cubic B-spline and the same knot vector with 50 bases.
# find basis functions
num_coeff = 50
clips = ['oceans','overcome']
pids = [1,2]
bases = np.empty((len(clips),len(pids)), dtype=object)
info = np.empty((len(clips),len(pids)), dtype=object)
for clip, clip_name in enumerate(clips):
for pid_i, pid in enumerate(pids):
temp_df = traj_df[(traj_df.clip_name==clip_name) & (traj_df.pid==pid)]
data = temp_df[['x','y','z']].to_numpy()
tck, u = interpolate.splprep(data.T, k=3, u=np.linspace(0,1,temp_df['clip_len'].iloc[0]), t=np.concatenate((np.array([0,0,0]),np.linspace(0,1,num_coeff-2),np.array([1,1,1]))), task=-1)
temp_df['clip_len'] = temp_df['clip_len']-1
t_sym = symbols('t')
basis = bspline_basis_set(tck[2], tck[0].tolist(), t_sym)
bases[clip][pid_i] = basis
info[clip][pid_i] = clip_name+' '+str(pid)
# print basis functions:
bases_to_display = [0,1,-1,len(bases[0][0])-2,len(bases[0][0])-1]
for basis in bases_to_display:
if (basis==-1):
print('...')
else:
print(f'Basis {basis+1}:')
for clip in range(len(bases)):
for pid_i in range(len(bases[0])):
basis_str = ''
for piece in range(len(bases[clip][pid_i][basis].args)-1):
basis_str += str((bases[clip][pid_i][basis].args[piece])[1]) # t
basis_str += '\n'+' '*17
basis_str += str((bases[clip][pid_i][basis].args[piece])[0]) # basis function
basis_str += '\n'+' '*13
print(f'{info[clip][pid_i]:10s} : {basis_str}')
print()
Basis 1:
oceans 1 : (t >= 0) & (t <= 0.0212765957446809)
-103823.0*t**3 + 6627.0*t**2 - 141.0*t + 1.0
oceans 2 : (t >= 0) & (t <= 0.0212765957446809)
-103823.0*t**3 + 6627.0*t**2 - 141.0*t + 1.0
overcome 1 : (t >= 0) & (t <= 0.0212765957446809)
-103823.0*t**3 + 6627.0*t**2 - 141.0*t + 1.0
overcome 2 : (t >= 0) & (t <= 0.0212765957446809)
-103823.0*t**3 + 6627.0*t**2 - 141.0*t + 1.0
Basis 2:
oceans 1 : (t >= 0) & (t <= 0.0212765957446809)
181690.25*t**3 - 9940.5*t**2 + 141.0*t
(t >= 0.0212765957446809) & (t <= 0.0425531914893617)
-25955.75*t**3 + 3313.5*t**2 - 141.0*t + 2.0
oceans 2 : (t >= 0) & (t <= 0.0212765957446809)
181690.25*t**3 - 9940.5*t**2 + 141.0*t
(t >= 0.0212765957446809) & (t <= 0.0425531914893617)
-25955.75*t**3 + 3313.5*t**2 - 141.0*t + 2.0
overcome 1 : (t >= 0) & (t <= 0.0212765957446809)
181690.25*t**3 - 9940.5*t**2 + 141.0*t
(t >= 0.0212765957446809) & (t <= 0.0425531914893617)
-25955.75*t**3 + 3313.5*t**2 - 141.0*t + 2.0
overcome 2 : (t >= 0) & (t <= 0.0212765957446809)
181690.25*t**3 - 9940.5*t**2 + 141.0*t
(t >= 0.0212765957446809) & (t <= 0.0425531914893617)
-25955.75*t**3 + 3313.5*t**2 - 141.0*t + 2.0
...
Basis 49:
oceans 1 : (t >= 0.957446808510638) & (t <= 0.978723404255319)
25955.7499999999*t**3 - 74553.7499999997*t**2 + 71381.2499999997*t - 22781.2499999999
(t >= 0.978723404255319) & (t <= 1.0)
-181690.249999999*t**3 + 535130.249999997*t**2 - 525330.749999998*t + 171890.749999999
oceans 2 : (t >= 0.957446808510638) & (t <= 0.978723404255319)
25955.7499999999*t**3 - 74553.7499999997*t**2 + 71381.2499999997*t - 22781.2499999999
(t >= 0.978723404255319) & (t <= 1.0)
-181690.249999999*t**3 + 535130.249999997*t**2 - 525330.749999998*t + 171890.749999999
overcome 1 : (t >= 0.957446808510638) & (t <= 0.978723404255319)
25955.7499999999*t**3 - 74553.7499999997*t**2 + 71381.2499999997*t - 22781.2499999999
(t >= 0.978723404255319) & (t <= 1.0)
-181690.249999999*t**3 + 535130.249999997*t**2 - 525330.749999998*t + 171890.749999999
overcome 2 : (t >= 0.957446808510638) & (t <= 0.978723404255319)
25955.7499999999*t**3 - 74553.7499999997*t**2 + 71381.2499999997*t - 22781.2499999999
(t >= 0.978723404255319) & (t <= 1.0)
-181690.249999999*t**3 + 535130.249999997*t**2 - 525330.749999998*t + 171890.749999999
Basis 50:
oceans 1 : (t >= 0.978723404255319) & (t <= 1.0)
103823.0*t**3 - 304841.999999999*t**2 + 298355.999999999*t - 97335.9999999995
oceans 2 : (t >= 0.978723404255319) & (t <= 1.0)
103823.0*t**3 - 304841.999999999*t**2 + 298355.999999999*t - 97335.9999999995
overcome 1 : (t >= 0.978723404255319) & (t <= 1.0)
103823.0*t**3 - 304841.999999999*t**2 + 298355.999999999*t - 97335.9999999995
overcome 2 : (t >= 0.978723404255319) & (t <= 1.0)
103823.0*t**3 - 304841.999999999*t**2 + 298355.999999999*t - 97335.9999999995
From these examples, we can see that the bases are identical regardless of the clip or participant, which is consistent with the definition of a B-spline. We can also notice that they are sorted by \(u\), which corresponds to time in the trajectory.
We can plot these basis functions, though we’ll only use 10 bases for a better visualization.
# find basis functions
num_bases = 10
temp_df = traj_df[(traj_df.clip_name=='oceans') & (traj_df.pid==1)]
data = temp_df[['x','y','z']].to_numpy()
tck, u = interpolate.splprep(data.T, k=3, u=np.linspace(0,1,temp_df['clip_len'].iloc[0]), t=np.concatenate((np.array([0,0,0]),np.linspace(0,1,num_bases-2),np.array([1,1,1]))), task=-1)
temp_df['clip_len'] = temp_df['clip_len']-1
t_sym = symbols('t')
bases = bspline_basis_set(tck[2], tck[0].tolist(), t_sym)
# plot bases
plotly_data = []
colorscale = np.array(colors)
colorscale = np.tile(colors, num_bases//len(colors)+1)
t_between = 1/(num_bases-3)
for basis in range(1,num_bases+1):
t_low = (basis-4)*t_between
t_high = (basis)*t_between
if (t_low < 0):
t_low = 0
if (t_high > 1):
t_high = 1
t_eval = np.linspace(t_low,t_high,int((t_high-t_low)/t_between)*50)
x = np.zeros(len(t_eval))
for i,t in enumerate(t_eval):
x[i] = bases[basis-1].evalf(subs={t_sym:t})
basis_func = go.Scatter(
x=t_eval*(num_bases-3),
y=x,
mode='lines',
customdata=np.round(np.vstack((np.ones(len(t_eval))*(t_eval[0]*(num_bases-3)), np.ones(len(t_eval))*(t_eval[-1]*(num_bases-3)))).T),
hovertemplate='[%{customdata[0]},%{customdata[1]}]',
line={'width':4, 'color':colorscale[basis-1]},
name='Basis '+str(basis),
showlegend=False
)
plotly_data.append(basis_func)
# formatting
plotly_layout = go.Layout(
autosize=False,
showlegend=True,
width=800,
height=600,
margin={'l':0, 'r':0, 't':40, 'b':60},
legend={'orientation':'h',
'itemsizing':'constant',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.07,
'tracegroupgap':2},
title={'text':'B-Spline Basis Functions',
'xanchor':'center',
'yanchor':'top',
'x':0.5,
'y':0.98},
hovermode='closest',
annotations=[{'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.11,
'showarrow':False,
'text':'<b>Fig. 1.</b> B-spline basis functions, with '+str(num_bases)+' bases in total. Time t is scaled to represent segments between adjacent knots.'}])
plotly_config = {'displaylogo':False,
'modeBarButtonsToRemove': ['autoScale2d','toggleSpikelines','hoverClosestCartesian','hoverCompareCartesian','lasso2d','select2d']}
fig = go.Figure(data=plotly_data, layout=plotly_layout)
fig.show(config=plotly_config)
Coefficients¶
Now we can calculate the coefficients for each trajectory. Since our trajectories are in 3-space, there will be a coefficient for each dimension (x,y,z) that corresponds to each basis.
num_coeff = 50
c_x = np.empty(0)
c_y = np.empty(0)
c_z = np.empty(0)
basis = np.empty(0,dtype=int)
for clip, clip_name in enumerate(clipdata_df['clip_name']):
temp_df = traj_df[(traj_df.clip_name==clip_name)]
for pid in range(max(temp_df.pid)):
data = temp_df[temp_df.pid==pid+1][['x','y','z']].to_numpy()
tck, u = interpolate.splprep(data.T, k=3, u=np.linspace(0,1,temp_df['clip_len'].iloc[0]), t=np.concatenate((np.array([0,0,0]),np.linspace(0,1,num_coeff-2),np.array([1,1,1]))), task=-1)
c_x = np.append(c_x, tck[1][0])
c_y = np.append(c_y, tck[1][1])
c_z = np.append(c_z, tck[1][2])
basis = np.append(basis, np.arange(0,num_coeff,dtype=int))
temp_df = temp_df[temp_df.time==1].drop(columns=['time'])
coeff_df = traj_df[traj_df.time==1].drop(columns=['time'])
coeff_df = coeff_df.iloc[np.arange(len(coeff_df)).repeat(num_coeff)] # duplicate rows
coeff_df['basis'] = basis
coeff_df['x'] = c_x
coeff_df['y'] = c_y
coeff_df['z'] = c_z
coeff_df = coeff_df[['clip','clip_name','clip_len','pid','basis','x','y','z']] # reorder columns
coeff_df = coeff_df.rename(columns={'x': 'c_x', 'y': 'c_y', 'z': 'c_z'})
coeff_df = coeff_df.reset_index(drop=True)
display(coeff_df)
| clip | clip_name | clip_len | pid | basis | c_x | c_y | c_z | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0 | testretest | 84 | 1 | 0 | -0.071108 | 0.290005 | 0.076626 |
| 1 | 0 | testretest | 84 | 1 | 1 | -1.164143 | 1.108597 | 0.882055 |
| 2 | 0 | testretest | 84 | 1 | 2 | 0.446077 | -1.231524 | -1.698370 |
| 3 | 0 | testretest | 84 | 1 | 3 | 0.195367 | 1.690015 | 3.639953 |
| 4 | 0 | testretest | 84 | 1 | 4 | -1.389411 | -2.023206 | 0.279725 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 68395 | 14 | starwars | 256 | 76 | 45 | -2.362159 | -18.256953 | 17.115972 |
| 68396 | 14 | starwars | 256 | 76 | 46 | -6.569614 | -17.539154 | 17.910120 |
| 68397 | 14 | starwars | 256 | 76 | 47 | -6.084877 | -13.444515 | 9.311504 |
| 68398 | 14 | starwars | 256 | 76 | 48 | -5.553723 | -15.126896 | 17.265603 |
| 68399 | 14 | starwars | 256 | 76 | 49 | -3.707036 | -13.710217 | 16.163879 |
68400 rows × 8 columns
Before classification, we can take a look at the coefficients.
fig = make_subplots(rows=3, cols=2,
shared_xaxes=True,
vertical_spacing=0.03, horizontal_spacing=0.05,
subplot_titles=('x','x','y','y','z','z'),
specs=[[{'type':'scatter'}, {'type':'scatter'}], [{'type':'scatter'}, {'type':'scatter'}], [{'type':'scatter'}, {'type':'scatter'}]])
for clip, clip_name in enumerate(clipdata_df['clip_name']):
# smoothed (splines)
temp_df = coeff_df[(coeff_df.clip_name==clip_name)]
temp_df['mean_c_x'] = temp_df.groupby('basis')['c_x'].transform('mean')
temp_df['std_c_x'] = temp_df.groupby('basis')['c_x'].transform('std')
temp_df['mean_c_y'] = temp_df.groupby('basis')['c_y'].transform('mean')
temp_df['std_c_y'] = temp_df.groupby('basis')['c_y'].transform('std')
temp_df['mean_c_z'] = temp_df.groupby('basis')['c_z'].transform('mean')
temp_df['std_c_z'] = temp_df.groupby('basis')['c_z'].transform('std')
temp_df = temp_df[temp_df.pid==1]
# visibility = 'legendonly'
# if (clip_name=='oceans'):
# visibility = True
visibility = True
for row, var in enumerate(['x','y','z']):
row += 1
mean = 'mean_c_'+var
std = 'std_c_'+var
if (var=='x'):
showlegend=True
else:
showlegend=False
# c (no std)
mean_traj = go.Scatter(
x=temp_df['basis']+1,
y=temp_df[mean],
customdata=temp_df[std],
mode='markers+lines',
line={'width':2, 'color':colors[clip]},
marker={'size':4, 'color':colors[clip]},
name=clip_name,
legendgroup=clip_name,
showlegend=showlegend,
visible=visibility,
hovertemplate='basis: %{x}<br>coeff: %{y:.3f}<br>sd: %{customdata:.3f}'
)
fig.add_trace(mean_traj, row=row, col=1)
# c (std)
mean_traj = go.Scatter(
x=temp_df['basis']+1,
y=temp_df[mean],
customdata=temp_df[std],
mode='markers+lines',
line={'width':2, 'color':colors[clip]},
marker={'size':4, 'color':colors[clip]},
name=clip_name,
legendgroup=clip_name,
showlegend=False,
visible=visibility,
hovertemplate='basis: %{x}<br>coeff: %{y:.3f}<br>sd: %{customdata:.3f}'
)
fig.add_trace(mean_traj, row=row, col=2)
upper = temp_df[mean] + temp_df[std]
lower = temp_df[mean] - temp_df[std]
std_traj = go.Scatter(
x=np.concatenate([temp_df.index, temp_df.index[::-1]])-temp_df.index[0]+1,
y=pd.concat([upper, lower[::-1]]),
fill='toself',
mode='lines',
line={'width':0, 'color':colors[clip]},
opacity=0.7,
name=clip_name,
legendgroup=clip_name,
showlegend=False,
visible=visibility,
hoverinfo='skip'
)
fig.add_trace(std_traj, row=row, col=2)
# formatting
fig.update_layout(
autosize=False,
showlegend=True,
width=800,
height=1200,
margin={'l':0, 'r':0, 't':70, 'b':120},
legend={'orientation':'h',
'itemsizing':'constant',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.07,
'tracegroupgap':2},
title={'text':'Mean Individual Trajectory B-Spline Basis Function Coefficients',
'xanchor':'center',
'yanchor':'top',
'x':0.5,
'y':0.98},
hovermode='closest')
fig['layout']['annotations'] += (
{'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.12,
'showarrow':False,
'text':'<b>Fig. 2.</b> Mean basis function coefficients across all participants for each clip.<br>Error bars show the standard deviation of the mean basis coefficients.'
},
)
plotly_config = {'displaylogo':False,
'modeBarButtonsToRemove': ['autoScale2d','toggleSpikelines','hoverClosestCartesian','hoverCompareCartesian','lasso2d','select2d']}
fig.show(config=plotly_config)
These coefficients look reasonably separable, so it seems that classification seems possible. Each clip’s mean coefficients are fairly distanced from other clips. The standard deviation isn’t too high, which indicates a lower variability that would make it easier to consistently predict clips. However, there is some overlap between clips.
Classification¶
Dataset¶
Before beginning classification, we want to scale our features using mean normalization:
This restricts all features between -1 and 1 as well as setting the mean of each feature to 0.
for c in ['c_x','c_y','c_z']:
coeff_df[c] = (coeff_df[c] - coeff_df[c].mean()) / (max(coeff_df[c]) - min(coeff_df[c]))
display(coeff_df)
| clip | clip_name | clip_len | pid | basis | c_x | c_y | c_z | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0 | testretest | 84 | 1 | 0 | -0.024249 | 0.021235 | -0.043516 |
| 1 | 0 | testretest | 84 | 1 | 1 | -0.045517 | 0.033720 | -0.029166 |
| 2 | 0 | testretest | 84 | 1 | 2 | -0.014185 | -0.001971 | -0.075140 |
| 3 | 0 | testretest | 84 | 1 | 3 | -0.019064 | 0.042588 | 0.019969 |
| 4 | 0 | testretest | 84 | 1 | 4 | -0.049901 | -0.014046 | -0.039897 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 68395 | 14 | starwars | 256 | 76 | 45 | -0.068829 | -0.261642 | 0.260062 |
| 68396 | 14 | starwars | 256 | 76 | 46 | -0.150698 | -0.250694 | 0.274211 |
| 68397 | 14 | starwars | 256 | 76 | 47 | -0.141266 | -0.188243 | 0.121015 |
| 68398 | 14 | starwars | 256 | 76 | 48 | -0.130931 | -0.213902 | 0.262728 |
| 68399 | 14 | starwars | 256 | 76 | 49 | -0.094997 | -0.192295 | 0.243099 |
68400 rows × 8 columns
To represent a trajectory as a single input to a model, we can combine the x, y, and z coefficients across all bases for a single participant and clip. This yields a coefficient array of length 150 (50 bases * 3 dimensions) for each participant/clip combination. Essentially, the ‘c’ column represents a 2d array of coefficients, which is also stored in a numpy array for convenience.
num_coeff = max(coeff_df['basis'])+1
coeff_np = np.empty((0,num_coeff*3))
for traj in range(len(coeff_df)//num_coeff):
temp_c = np.zeros(0)
for basis in range(num_coeff):
i = traj*num_coeff + basis
temp_c = np.append(temp_c, [coeff_df['c_x'].iloc[i], coeff_df['c_y'].iloc[i], coeff_df['c_z'].iloc[i]])
coeff_np = np.vstack((coeff_np,temp_c))
coeff_df = coeff_df[coeff_df.basis==0].drop(columns=['basis','c_y','c_z'])
clip_np = coeff_df['clip'].to_numpy()
coeff_df = coeff_df.astype(object)
for i in range(len(coeff_df)):
coeff_df['c_x'].iloc[i] = coeff_np[i]
coeff_df = coeff_df.rename(columns={'c_x': 'c'})
coeff_df = coeff_df.reset_index(drop=True)
display(coeff_df)
| clip | clip_name | clip_len | pid | c | |
|---|---|---|---|---|---|
| 0 | 0 | testretest | 84 | 1 | [-0.024248835663933252, 0.021235331253160083, ... |
| 1 | 0 | testretest | 84 | 2 | [-0.03599982880421413, 0.009646510915489412, -... |
| 2 | 0 | testretest | 84 | 3 | [0.0027464221244179526, 0.004096925643923165, ... |
| 3 | 0 | testretest | 84 | 4 | [-0.04091597762027432, -0.011983818806115683, ... |
| 4 | 0 | testretest | 84 | 5 | [-0.044545932726389674, 0.001223617590038054, ... |
| ... | ... | ... | ... | ... | ... |
| 1363 | 14 | starwars | 256 | 72 | [-0.039080858100526895, 0.019357595602992807, ... |
| 1364 | 14 | starwars | 256 | 73 | [-0.005983522682653093, -0.030670381289564837,... |
| 1365 | 14 | starwars | 256 | 74 | [-0.03055848267077042, 0.018508536322194236, -... |
| 1366 | 14 | starwars | 256 | 75 | [-0.01903681855484776, 0.04047392725180849, -0... |
| 1367 | 14 | starwars | 256 | 76 | [-0.038507534109979605, -0.009752887872791728,... |
1368 rows × 5 columns
# create dataset
coefficients = torch.from_numpy(coeff_np.astype(np.float32))
clips = torch.from_numpy(clip_np.astype(np.int))
coeff_dataset = TensorDataset(coefficients, clips)
Having reduced each trajectory to coefficients, we can take try classifying clips by providing a model with coefficients rather than spatial-temporal information about the trajectory itself.
Multilayer Perceptron (MLP)¶
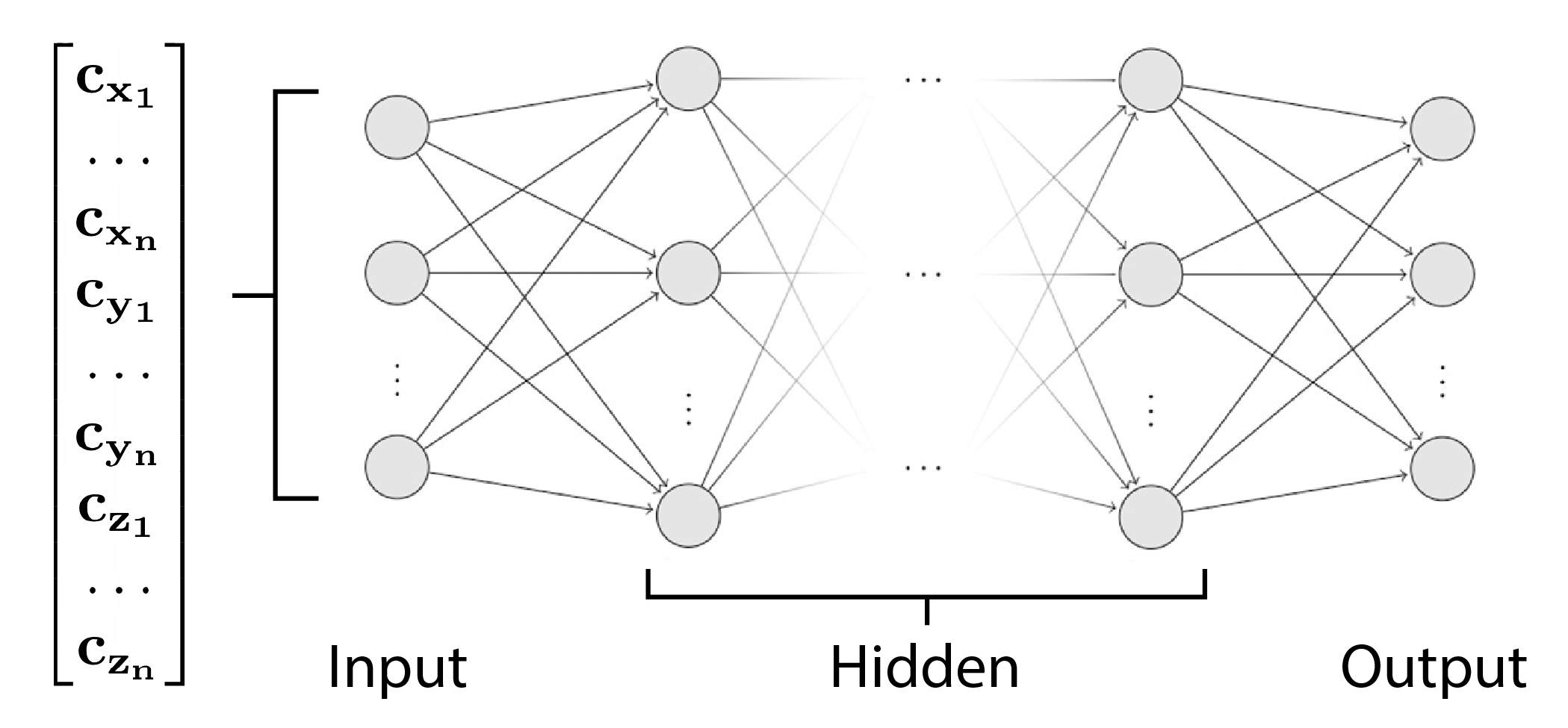 We can begin with a simple MLP model with one hidden layer for classification. The model will recieve all coefficients as input. This comes in the form of 50 coefficients for each dimension, totalling to 150 coefficients.
We can begin with a simple MLP model with one hidden layer for classification. The model will recieve all coefficients as input. This comes in the form of 50 coefficients for each dimension, totalling to 150 coefficients.
To estimate the model’s generalization performance, we’ll use 10-fold cross validation using a grid search.
class MLP(nn.Module):
def __init__(self, k_input, k_hidden, k_layers, k_output, dropout=0):
nn.Module.__init__(self) #super().__init__()
self.k_layers = k_layers
self.fc = nn.ModuleList([])
for i in range(k_layers+2):
if (i==0):
self.fc.append(nn.Linear(k_input, k_hidden))
elif (i==k_layers+1):
self.fc.append(nn.Linear(k_hidden, k_output))
else:
self.fc.append(nn.Linear(k_hidden, k_hidden))
self.dropout = nn.Dropout(dropout)
def forward(self, x):
for i in range(self.k_layers+1):
x = self.dropout(F.relu(self.fc[i](x)))
y = self.fc[i+1](x)
return y
def train(model, criterion, optimizer, dataloader, device):
model.train()
model = model.to(device)
criterion = criterion.to(device)
running_loss = 0.0
running_corrects = 0.0
for inputs, labels in dataloader:
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
loss = criterion(outputs, labels)
optimizer.zero_grad()
loss.backward()
optimizer.step()
_, preds = torch.max(outputs, 1)
running_loss += loss.detach() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
epoch_loss = running_loss / len(dataloader.dataset)
epoch_acc = running_corrects.float() / len(dataloader.dataset)
return epoch_loss, epoch_acc
def evaluate(model, criterion, dataloader, device):
model.eval()
model = model.to(device)
criterion = criterion.to(device)
running_loss = 0.0
running_corrects = 0.0
with torch.no_grad():
for inputs, labels in dataloader:
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
loss = criterion(outputs, labels)
_, preds = torch.max(outputs, 1)
running_loss += loss.detach() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
epoch_loss = running_loss / len(dataloader.dataset)
epoch_acc = running_corrects.float() / len(dataloader.dataset)
return epoch_loss, epoch_acc
# evaluate and return a confusion matrix
def evaluate_cm(model, criterion, dataloader, device):
model.eval()
model = model.to(device)
criterion = criterion.to(device)
running_loss = 0.0
running_corrects = 0.0
y_true = np.zeros(0,dtype=np.int)
y_pred = np.zeros(0,dtype=np.int)
with torch.no_grad():
for inputs, labels in dataloader:
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
loss = criterion(outputs, labels)
_, preds = torch.max(outputs, 1)
running_loss += loss.detach() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
y_true = np.append(y_true, labels.cpu().numpy())
y_pred = np.append(y_pred, preds.cpu().numpy())
epoch_loss = running_loss / len(dataloader.dataset)
epoch_acc = running_corrects.float() / len(dataloader.dataset)
# confusion matrix
cm = confusion_matrix(y_true, y_pred)
return epoch_loss, epoch_acc, cm
def grid_search_mlp(train_dl, val_dl, num_epochs, k_hidden, k_layers, scoring, criterion, device, dropout=0):
val_scores = np.zeros((len(k_hidden),len(k_layers)))
for i,hidden in enumerate(k_hidden):
for j,layers in enumerate(k_layers):
model = MLP(150,hidden,layers,15,dropout)
optimizer = optim.Adam(model.parameters())
avg_val_score = 0
for epoch in range(num_epochs-5):
train(model, criterion, optimizer, train_dl, device)
for epoch in range(5):
train(model, criterion, optimizer, train_dl, device)
val_loss, val_acc = evaluate(model, criterion, val_dl, device)
# if (scoring=='acc' or scoring=='accuracy'):
# avg_val_score += val_acc
# elif (scoring=='loss'):
# avg_val_score += val_loss
avg_val_score += val_acc # omit if statement for speed
avg_val_score /= 5
val_scores[i][j] = avg_val_score
return val_scores
def nested_cv_mlp(dataset, outer_kfold, num_outer_epochs, inner_kfold, num_inner_epochs, batch_size, k_hidden, k_layers, k_seed, scoring, device, opt_title, opt_caption, lossacc_title, lossacc_caption, cm_title, cm_caption, dropout=0):
# set seed for reproducibility
torch.manual_seed(k_seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
# NESTED CV
loss_acc_df = pd.DataFrame(columns=['fold','epoch','train_loss','train_acc','test_loss','test_acc'])
x, y = dataset[:]
cm = np.zeros((torch.max(y).item()+1,torch.max(y).item()+1), dtype=np.int)
total_val_score = np.zeros((len(k_hidden),len(k_layers)))
total_freq = np.zeros((len(k_hidden),len(k_layers)), dtype=np.int)
outer_kfold = KFold(n_splits=outer_kfold, shuffle=True, random_state=0)
inner_kfold = KFold(n_splits=inner_kfold, shuffle=True, random_state=0)
criterion = nn.CrossEntropyLoss()
# Outer CV (trainval/test split)
current_outer_fold = 1
for trainval_index, test_index in outer_kfold.split(x, y):
print(f'Outer fold {current_outer_fold}/{outer_kfold.n_splits}')
x_trainval = x[trainval_index]
y_trainval = y[trainval_index]
x_test = x[test_index]
y_test = y[test_index]
trainval_data = TensorDataset(x_trainval, y_trainval)
test_data = TensorDataset(x_test, y_test)
# Inner CV (train/val split)
current_inner_fold = 1
inner_val_score = np.zeros((len(k_hidden),len(k_layers)))
for train_index, val_index in inner_kfold.split(x_trainval, y_trainval):
print(f' Inner fold {current_inner_fold}/{inner_kfold.n_splits}')
train_data = TensorDataset(x_trainval[train_index], y_trainval[train_index])
val_data = TensorDataset(x_trainval[val_index], y_trainval[val_index])
train_dl = DataLoader(train_data, batch_size=batch_size, shuffle=True)
val_dl = DataLoader(val_data, batch_size=batch_size, shuffle=True)
fold_val_score = grid_search_mlp(train_dl, val_dl, num_inner_epochs, k_hidden, k_layers, scoring, criterion, device, dropout)
inner_val_score = np.add(inner_val_score, fold_val_score)
current_inner_fold += 1
if (scoring=='acc' or scoring=='accuracy'):
best_params = np.unravel_index(np.argmax(inner_val_score, axis=None), inner_val_score.shape)
elif (scoring=='loss'):
best_params = np.unravel_index(np.argmin(inner_val_score, axis=None), inner_val_score.shape)
total_freq[best_params[0],best_params[1]] += 1
model = MLP(150,k_hidden[best_params[0]],k_layers[best_params[1]],15,dropout)
optimizer = optim.Adam(model.parameters())
trainval_dl = DataLoader(trainval_data, batch_size=batch_size, shuffle=True)
test_dl = DataLoader(test_data, batch_size=batch_size, shuffle=True)
for epoch in range(1,num_outer_epochs+1):
trainval_loss, trainval_acc = train(model, criterion, optimizer, trainval_dl, device)
if (epoch==num_outer_epochs):
test_loss, test_acc, fold_cm = evaluate_cm(model, criterion, test_dl, device)
cm += fold_cm
else:
test_loss, test_acc = evaluate(model, criterion, test_dl, device)
loss_acc_df = loss_acc_df.append({
'fold':current_outer_fold,
'epoch':epoch,
'train_loss':trainval_loss.item(),
'train_acc':trainval_acc.item(),
'test_loss':test_loss.item(),
'test_acc':test_acc.item()},
ignore_index=True)
current_outer_fold += 1
total_val_score = np.add(total_val_score, inner_val_score)
# PLOT HYPERPARAMETERS
clear_output()
avg_val_score = total_val_score / (outer_kfold.n_splits * inner_kfold.n_splits)
avg_freq = total_freq / outer_kfold.n_splits
plotly_data = []
plotly_data.append(
go.Heatmap(z=np.flip(avg_val_score,axis=0), y=np.flip(np.array(k_hidden)), x=k_layers, colorscale='blues',
zmin=np.amin(avg_val_score), zmax=np.amax(avg_val_score),
name='Validation<br>Accuracy',
customdata=np.flip(avg_freq,axis=0),
hovertemplate='k_hidden: %{y}<br>k_layers: %{x}<br>'+scoring+': %{z:.5f}<br>rel freq: %{customdata:.3f}',
visible=True))
plotly_data.append(
go.Heatmap(z=np.flip(avg_freq,axis=0), y=np.flip(np.array(k_hidden)), x=k_layers, colorscale='blues',
zmin=np.amin(avg_freq), zmax=np.amax(avg_freq),
name='Relative<br>Frequency',
customdata=np.flip(avg_val_score,axis=0),
hovertemplate='k_hidden: %{y}<br>k_layers: %{x}<br>'+scoring+': %{customdata:.5f}<br>rel freq: %{z:.3f}',
visible=False))
acc_annotations = []
mid = (np.amin(avg_val_score)+np.amax(avg_val_score))/2
for i, row in enumerate(avg_val_score.T):
for j, value in enumerate(row):
if (value > mid):
color = 'white'
else:
color = 'black'
acc_annotations.append({
'x': k_layers[i],
'y': k_hidden[j],
'font': {'color': color},
'text': str(round(value,5)),
'xref': 'x1',
'yref': 'y1',
'showarrow': False
})
acc_annotations.append({
'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.14,
'showarrow':False,
'text':opt_caption
})
freq_annotations = []
mid = (np.amin(avg_freq)+np.amax(avg_freq))/2
for i, row in enumerate(avg_freq.T):
for j, value in enumerate(row):
if (value > mid):
color = 'white'
else:
color = 'black'
freq_annotations.append({
'x': k_layers[i],
'y': k_hidden[j],
'font': {'color': color},
'text': str(round(value,3)),
'xref': 'x1',
'yref': 'y1',
'showarrow': False
})
freq_annotations.append({
'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.14,
'showarrow':False,
'text':opt_caption
})
plotly_layout = go.Layout(
autosize=False,
width=800,
height=800,
margin={'l':0, 'r':0, 't':40, 'b':100},
xaxis={'title': 'k_layers', 'fixedrange':True, 'type':'category'},
yaxis={'title': 'k_hidden', 'fixedrange':True, 'type':'category'},
title={'text':opt_title,
'xanchor':'center',
'yanchor':'top',
'x':0.5,
'y':0.98},
annotations=acc_annotations,
updatemenus=[{'type':'buttons',
'direction':'left',
'pad':{'l':0, 'r':0, 't':0, 'b':0},
'xanchor':'left',
'yanchor':'top',
'x':0,
'y':1.055,
'buttons':[
{'label':'Val. Acc.',
'method': 'update',
'args':[{'visible': [True,False]},
{'annotations': acc_annotations}]},
{'label':'Rel. Freq.',
'method': 'update',
'args':[{'visible': [False,True]},
{'annotations': freq_annotations}]}
]}])
plotly_config = {'displaylogo':False,
'modeBarButtonsToRemove': ['autoScale2d','toggleSpikelines','hoverClosestCartesian','hoverCompareCartesian','lasso2d','select2d','zoom2d','pan2d','zoomIn2d','zoomOut2d','resetScale2d']}
fig = go.Figure(data=plotly_data, layout=plotly_layout)
fig.show(config=plotly_config)
# PLOT LOSS / ACCURACY
loss_acc_df['mean_train_loss'] = loss_acc_df.groupby('epoch')['train_loss'].transform('mean')
loss_acc_df['std_train_loss'] = loss_acc_df.groupby('epoch')['train_loss'].transform('std')
loss_acc_df['mean_train_acc'] = loss_acc_df.groupby('epoch')['train_acc'].transform('mean')
loss_acc_df['std_train_acc'] = loss_acc_df.groupby('epoch')['train_acc'].transform('std')
loss_acc_df['mean_test_loss'] = loss_acc_df.groupby('epoch')['test_loss'].transform('mean')
loss_acc_df['std_test_loss'] = loss_acc_df.groupby('epoch')['test_loss'].transform('std')
loss_acc_df['mean_test_acc'] = loss_acc_df.groupby('epoch')['test_acc'].transform('mean')
loss_acc_df['std_test_acc'] = loss_acc_df.groupby('epoch')['test_acc'].transform('std')
loss_acc_df = loss_acc_df[loss_acc_df.fold==1]
fig = make_subplots(rows=2, cols=1,
shared_xaxes=True,
vertical_spacing=0.05,
subplot_titles=('Loss','Accuracy'),
specs=[[{'type':'scatter'}], [{'type':'scatter'}]])
for dataset in ['train','test']:
if (dataset=='train'):
color = 'mediumblue'
elif (dataset=='test'):
color = 'crimson'
# loss (no std)
loss = go.Scatter(
x=loss_acc_df['epoch'],
y=loss_acc_df['mean_'+dataset+'_loss'],
customdata=loss_acc_df['std_'+dataset+'_loss'],
mode='markers+lines',
line={'width':2, 'color':color},
marker={'size':4, 'color':color},
name=dataset,
legendgroup=dataset,
showlegend=True,
visible=True,
hovertemplate='epoch: %{x}<br>loss: %{y:.5f}<br>sd: %{customdata:.5f}'
)
fig.add_trace(loss, row=1, col=1)
# loss (std)
upper = loss_acc_df['mean_'+dataset+'_loss'] + loss_acc_df['std_'+dataset+'_loss']
lower = loss_acc_df['mean_'+dataset+'_loss'] - loss_acc_df['std_'+dataset+'_loss']
loss = go.Scatter(
x=np.concatenate([loss_acc_df.index, loss_acc_df.index[::-1]])-loss_acc_df.index[0]+1,
y=pd.concat([upper, lower[::-1]]),
fill='toself',
mode='lines',
line={'width':0, 'color':color},
opacity=0.7,
name=dataset,
legendgroup=dataset,
showlegend=False,
visible=True,
hoverinfo='skip'
)
fig.add_trace(loss, row=1, col=1)
# acc (no std)
acc = go.Scatter(
x=loss_acc_df['epoch'],
y=loss_acc_df['mean_'+dataset+'_acc'],
customdata=loss_acc_df['std_'+dataset+'_acc'],
mode='markers+lines',
line={'width':2, 'color':color},
marker={'size':4, 'color':color},
name=dataset,
legendgroup=dataset,
showlegend=False,
visible=True,
hovertemplate='epoch: %{x}<br>acc: %{y:.5f}<br>sd: %{customdata:.5f}'
)
fig.add_trace(acc, row=2, col=1)
# acc (std)
upper = loss_acc_df['mean_'+dataset+'_acc'] + loss_acc_df['std_'+dataset+'_acc']
lower = loss_acc_df['mean_'+dataset+'_acc'] - loss_acc_df['std_'+dataset+'_acc']
acc = go.Scatter(
x=np.concatenate([loss_acc_df.index, loss_acc_df.index[::-1]])-loss_acc_df.index[0]+1,
y=pd.concat([upper, lower[::-1]]),
fill='toself',
mode='lines',
line={'width':0, 'color':color},
opacity=0.7,
name=dataset,
legendgroup=dataset,
showlegend=False,
visible=True,
hoverinfo='skip'
)
fig.add_trace(acc, row=2, col=1)
# formatting
fig.update_layout(
autosize=False,
width=800,
height=800,
margin={'l':0, 'r':0, 't':70, 'b':100},
legend={'orientation':'h',
'itemsizing':'constant',
'xanchor':'center',
'yanchor':'bottom',
'y':-0.07,
'x':0.5},
title={'text':lossacc_title,
'xanchor':'center',
'yanchor':'top',
'x':0.5,
'y':0.98},
hovermode='x')
fig['layout']['annotations'] += (
{'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.14,
'showarrow':False,
'text':lossacc_caption
},
)
plotly_config = {'displaylogo':False,
'modeBarButtonsToRemove': ['autoScale2d','toggleSpikelines','hoverClosestCartesian','hoverCompareCartesian','lasso2d','select2d']}
fig.show(config=plotly_config)
# PLOT CONFUSION MATRIX
cm = cm.astype('float32')
for i in range(len(cm)):
cm[i] = cm[i]/np.sum(cm[i]) # rows/columns total to 1
labels=clipdata_df['clip_name'].to_numpy()
plotly_data = go.Heatmap(z=np.rot90(cm.T), y=np.flip(labels), x=labels, colorscale="blues",
zmin=0, zmax=1,
hovertemplate='True: %{y}<br>Predicted: %{x}<br>p: %{z:.5f}')
annotations = []
for i, row in enumerate(cm.T):
for j, value in enumerate(row):
if (value > 0.5):
color = 'white'
else:
color = 'black'
annotations.append({
'x': labels[i],
'y': labels[j],
'font': {'color': color},
'text': str(round(value,3)),
'xref': 'x1',
'yref': 'y1',
'showarrow': False
})
annotations.append({
'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.18,
'showarrow':False,
'text':cm_caption
})
plotly_layout = go.Layout(
autosize=False,
width=800,
height=800,
margin={'l':0, 'r':0, 't':40, 'b':120},
xaxis={'title': 'Predicted', 'fixedrange':True, 'type':'category'},
yaxis={'title': 'True', 'fixedrange':True, 'type':'category'},
title={'text':cm_title,
'xanchor':'center',
'yanchor':'top',
'x':0.5,
'y':0.98},
annotations=annotations)
plotly_config = {'displaylogo':False,
'modeBarButtonsToRemove': ['autoScale2d','toggleSpikelines','hoverClosestCartesian','hoverCompareCartesian','lasso2d','select2d','zoom2d','pan2d','zoomIn2d','zoomOut2d','resetScale2d']}
fig = go.Figure(data=plotly_data, layout=plotly_layout)
fig.show(config=plotly_config)
nested_cv_mlp(
dataset=coeff_dataset,
outer_kfold=10,
num_outer_epochs=50,
inner_kfold=10,
num_inner_epochs=50,
batch_size=16,
k_hidden=[15,30,50,75,100,150],
k_layers=[1,2,3,4,5],
k_seed=330,
scoring='acc',
dropout=0.5,
device=torch.device('cpu'), #torch.device('cuda:0' if torch.cuda.is_available() else 'cpu'),
opt_title='MLP Hyperparameter Optimization',
opt_caption='<b>Fig. 3.</b> MLP hyperparameter optimization using 10-fold nested cross validation. (A) Mean validation accuracy.<br>(B) Relative frequency of being chosen as the best model from the inner CV.',
lossacc_title='MLP Loss and Accuracy',
lossacc_caption='<b>Fig. 4.</b> MLP mean loss and accuracy for train and test sets across outer folds of 10-fold nested cross validation.<br>Error bars show the standard deviation of the mean loss and accuracy at each epoch.',
cm_title='MLP Confusion Matrix',
cm_caption='<b>Fig. 5.</b> Mean confusion matrix for trained MLP model across outer folds of 10-fold nested cross validation.'
)
87% accuracy is extremely good for a 15-way classification, so we should be suspicious. To make sure the learned decision boundary isn’t by chance, we can try randomly shuffling the labels. This should disrupt the data’s natural class-separability and severely reduce classification accuracy.
# create dataset with shuffled labels
coefficients = torch.from_numpy(coeff_np.astype(np.float32))
clip_np_copy = np.copy(clip_np)
np.random.shuffle(clip_np_copy)
clips = torch.from_numpy(clip_np_copy.astype(np.int))
coeff_dataset_shuffled = TensorDataset(coefficients, clips)
nested_cv_mlp(
dataset=coeff_dataset_shuffled,
outer_kfold=10,
num_outer_epochs=50,
inner_kfold=10,
num_inner_epochs=50,
batch_size=16,
k_hidden=[150],
k_layers=[1,2,3],
k_seed=330,
scoring='acc',
dropout=0.5,
device=torch.device('cpu'), #torch.device('cuda:0' if torch.cuda.is_available() else 'cpu'),
opt_title='MLP Hyperparameter Optimization (Shuffled Labels)',
opt_caption='<b>Fig. 6.</b> MLP hyperparameter optimization using 10-fold nested cross validation with shuffled labels.<br>(A) Mean validation accuracy. (B) Relative frequency of being chosen as the best model from the inner CV.',
lossacc_title='MLP Loss and Accuracy (Shuffled Labels)',
lossacc_caption='<b>Fig. 7.</b> MLP mean loss and accuracy for train and test sets with shuffled labels across outer folds of<br>10-fold nested cross validation. Error bars show the standard deviation of the mean loss and accuracy at each epoch.',
cm_title='MLP Confusion Matrix (Shuffled Labels)',
cm_caption='<b>Fig. 8.</b> Mean confusion matrix for trained MLP model across outer folds of 10-fold nested cross validation with shuffled labels.'
)
The accuracy drops to 22% when the labels are shuffled. From the confusion matrix, we can see that the model is simply gussing “testretest” for every example, which occurs since “testretest” has 4 times the examples as any other clip. This behavior suggests that there are clear decision boundaries, so the 87% accuracy model is likely valid.
Long Short-Term Memory (LSTM)¶
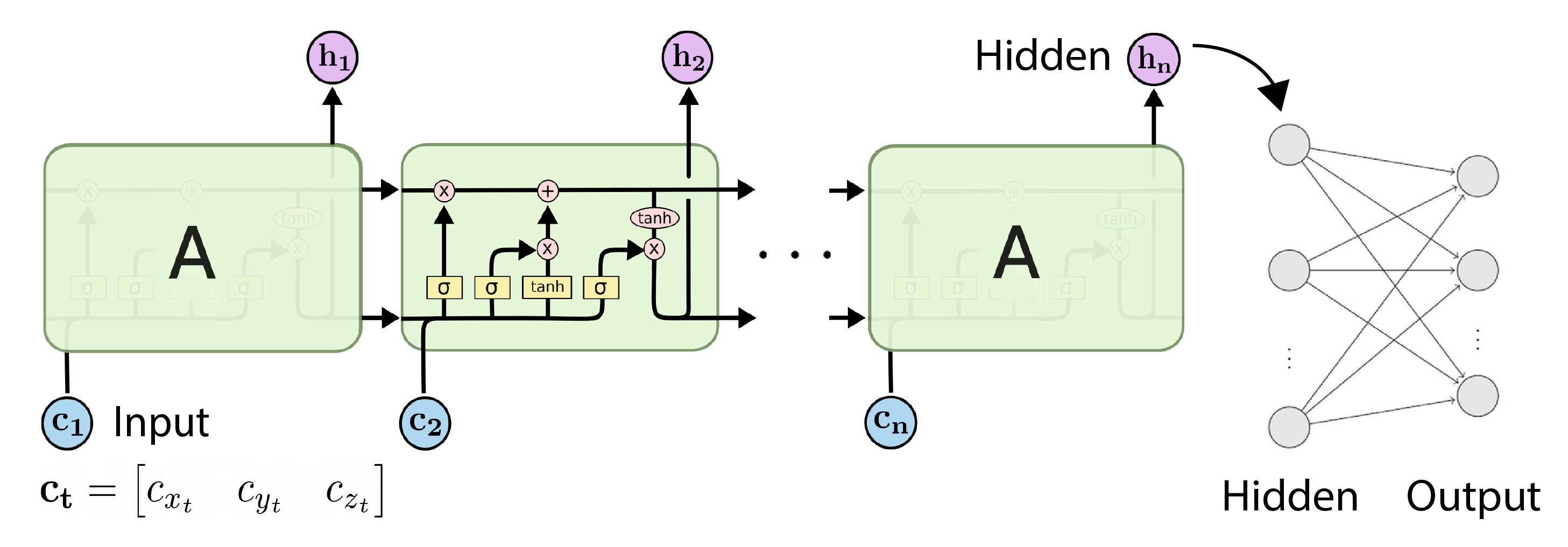
RNNs do have an advantage over MLPs since they can use information from hidden states, effectively creating a memory system that allows the RNN to use information from previous inputs rather than just the current input.
This is especially relevant with trajectories since they represent a state that is changing over time. This concept is retained when using B-splines. Each basis function contributes to a part of the B-spline defined by knots, which correspond to times in the trajectory. Since the basis functions are ordered with respect to time, we can also input their coefficients in order.
class LSTM(nn.Module):
def __init__(self, k_input, k_hidden, k_layers, k_output, dropout=0):
nn.Module.__init__(self) #super().__init__()
self.lstm = nn.LSTM(k_input, k_hidden, batch_first=True, num_layers=k_layers, dropout=dropout)
self.fc = nn.Linear(k_hidden, k_output)
def forward(self, x):
batch_size = x.shape[0]
x = x.view(batch_size,50,3) # (batch_size, seq_len, input_size)
out, (hidden, cell) = self.lstm(x)
y = self.fc(hidden[-1])
return y
def grid_search_lstm(train_dl, val_dl, num_epochs, k_hidden, k_layers, scoring, criterion, device, dropout):
val_scores = np.zeros((len(k_hidden),len(k_layers)))
for i,hidden in enumerate(k_hidden):
for j,layers in enumerate(k_layers):
model = LSTM(3,hidden,layers,15,dropout=dropout)
optimizer = optim.Adam(model.parameters())
avg_val_score = 0
for epoch in range(num_epochs-5):
train(model, criterion, optimizer, train_dl, device)
for epoch in range(5):
train(model, criterion, optimizer, train_dl, device)
val_loss, val_acc = evaluate(model, criterion, val_dl, device)
# if (scoring=='acc' or scoring=='accuracy'):
# avg_val_score += val_acc
# elif (scoring=='loss'):
# avg_val_score += val_loss
avg_val_score += val_acc # no if statement for speed
avg_val_score /= 5
val_scores[i][j] = avg_val_score
return val_scores
def nested_cv_lstm(dataset, outer_kfold, num_outer_epochs, inner_kfold, num_inner_epochs, batch_size, k_hidden, k_layers, k_seed, scoring, device, opt_title, opt_caption, lossacc_title, lossacc_caption, cm_title, cm_caption, dropout=0):
# set seed for reproducibility
torch.manual_seed(k_seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
# NESTED CV
loss_acc_df = pd.DataFrame(columns=['fold','epoch','train_loss','train_acc','test_loss','test_acc'])
x, y = dataset[:]
cm = np.zeros((torch.max(y).item()+1,torch.max(y).item()+1), dtype=np.int)
total_val_score = np.zeros((len(k_hidden),len(k_layers)))
total_freq = np.zeros((len(k_hidden),len(k_layers)), dtype=np.int)
outer_kfold = KFold(n_splits=outer_kfold, shuffle=True, random_state=0)
inner_kfold = KFold(n_splits=inner_kfold, shuffle=True, random_state=0)
criterion = nn.CrossEntropyLoss()
# Outer CV (trainval/test split)
current_outer_fold = 1
for trainval_index, test_index in outer_kfold.split(x, y):
print(f'Outer fold {current_outer_fold}/{outer_kfold.n_splits}')
x_trainval = x[trainval_index]
y_trainval = y[trainval_index]
x_test = x[test_index]
y_test = y[test_index]
trainval_data = TensorDataset(x_trainval, y_trainval)
test_data = TensorDataset(x_test, y_test)
# Inner CV (train/val split)
current_inner_fold = 1
inner_val_score = np.zeros((len(k_hidden),len(k_layers)))
for train_index, val_index in inner_kfold.split(x_trainval, y_trainval):
print(f' Inner fold {current_inner_fold}/{inner_kfold.n_splits}')
train_data = TensorDataset(x_trainval[train_index], y_trainval[train_index])
val_data = TensorDataset(x_trainval[val_index], y_trainval[val_index])
train_dl = DataLoader(train_data, batch_size=batch_size, shuffle=True)
val_dl = DataLoader(val_data, batch_size=batch_size, shuffle=True)
fold_val_score = grid_search_lstm(train_dl, val_dl, num_inner_epochs, k_hidden, k_layers, scoring, criterion, device, dropout)
inner_val_score = np.add(inner_val_score, fold_val_score)
current_inner_fold += 1
if (scoring=='acc' or scoring=='accuracy'):
best_params = np.unravel_index(np.argmax(inner_val_score, axis=None), inner_val_score.shape)
elif (scoring=='loss'):
best_params = np.unravel_index(np.argmin(inner_val_score, axis=None), inner_val_score.shape)
total_freq[best_params[0],best_params[1]] += 1
model = LSTM(3,k_hidden[best_params[0]],k_layers[best_params[1]],15,dropout)
optimizer = optim.Adam(model.parameters())
trainval_dl = DataLoader(trainval_data, batch_size=batch_size, shuffle=True)
test_dl = DataLoader(test_data, batch_size=batch_size, shuffle=True)
for epoch in range(1,num_outer_epochs+1):
trainval_loss, trainval_acc = train(model, criterion, optimizer, trainval_dl, device)
if (epoch==num_outer_epochs):
test_loss, test_acc, fold_cm = evaluate_cm(model, criterion, test_dl, device)
cm += fold_cm
else:
test_loss, test_acc = evaluate(model, criterion, test_dl, device)
loss_acc_df = loss_acc_df.append({
'fold':current_outer_fold,
'epoch':epoch,
'train_loss':trainval_loss.item(),
'train_acc':trainval_acc.item(),
'test_loss':test_loss.item(),
'test_acc':test_acc.item()},
ignore_index=True)
current_outer_fold += 1
total_val_score = np.add(total_val_score, inner_val_score)
# PLOT HYPERPARAMETERS
clear_output()
avg_val_score = total_val_score / (outer_kfold.n_splits * inner_kfold.n_splits)
avg_freq = total_freq / outer_kfold.n_splits
plotly_data = []
plotly_data.append(
go.Heatmap(z=np.flip(avg_val_score,axis=0), y=np.flip(np.array(k_hidden)), x=k_layers, colorscale='blues',
zmin=np.amin(avg_val_score), zmax=np.amax(avg_val_score),
name='Validation<br>Accuracy',
customdata=np.flip(avg_freq,axis=0),
hovertemplate='k_hidden: %{y}<br>k_layers: %{x}<br>'+scoring+': %{z:.5f}<br>rel freq: %{customdata:.3f}',
visible=True))
plotly_data.append(
go.Heatmap(z=np.flip(avg_freq,axis=0), y=np.flip(np.array(k_hidden)), x=k_layers, colorscale='blues',
zmin=np.amin(avg_freq), zmax=np.amax(avg_freq),
name='Relative<br>Frequency',
customdata=np.flip(avg_val_score,axis=0),
hovertemplate='k_hidden: %{y}<br>k_layers: %{x}<br>'+scoring+': %{customdata:.5f}<br>rel freq: %{z:.3f}',
visible=False))
acc_annotations = []
mid = (np.amin(avg_val_score)+np.amax(avg_val_score))/2
for i, row in enumerate(avg_val_score.T):
for j, value in enumerate(row):
if (value > mid):
color = 'white'
else:
color = 'black'
acc_annotations.append({
'x': k_layers[i],
'y': k_hidden[j],
'font': {'color': color},
'text': str(round(value,5)),
'xref': 'x1',
'yref': 'y1',
'showarrow': False
})
acc_annotations.append({
'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.14,
'showarrow':False,
'text':opt_caption
})
freq_annotations = []
mid = (np.amin(avg_freq)+np.amax(avg_freq))/2
for i, row in enumerate(avg_freq.T):
for j, value in enumerate(row):
if (value > mid):
color = 'white'
else:
color = 'black'
freq_annotations.append({
'x': k_layers[i],
'y': k_hidden[j],
'font': {'color': color},
'text': str(round(value,3)),
'xref': 'x1',
'yref': 'y1',
'showarrow': False
})
freq_annotations.append({
'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.14,
'showarrow':False,
'text':opt_caption
})
plotly_layout = go.Layout(
autosize=False,
width=800,
height=800,
margin={'l':0, 'r':0, 't':40, 'b':100},
xaxis={'title': 'k_layers', 'fixedrange':True, 'type':'category'},
yaxis={'title': 'k_hidden', 'fixedrange':True, 'type':'category'},
title={'text':opt_title,
'xanchor':'center',
'yanchor':'top',
'x':0.5,
'y':0.98},
annotations=acc_annotations,
updatemenus=[{'type':'buttons',
'direction':'left',
'pad':{'l':0, 'r':0, 't':0, 'b':0},
'xanchor':'left',
'yanchor':'top',
'x':0,
'y':1.055,
'buttons':[
{'label':'Val. Acc.',
'method': 'update',
'args':[{'visible': [True,False]},
{'annotations': acc_annotations}]},
{'label':'Rel. Freq.',
'method': 'update',
'args':[{'visible': [False,True]},
{'annotations': freq_annotations}]}
]}])
plotly_config = {'displaylogo':False,
'modeBarButtonsToRemove': ['autoScale2d','toggleSpikelines','hoverClosestCartesian','hoverCompareCartesian','lasso2d','select2d','zoom2d','pan2d','zoomIn2d','zoomOut2d','resetScale2d']}
fig = go.Figure(data=plotly_data, layout=plotly_layout)
fig.show(config=plotly_config)
# PLOT LOSS / ACCURACY
loss_acc_df['mean_train_loss'] = loss_acc_df.groupby('epoch')['train_loss'].transform('mean')
loss_acc_df['std_train_loss'] = loss_acc_df.groupby('epoch')['train_loss'].transform('std')
loss_acc_df['mean_train_acc'] = loss_acc_df.groupby('epoch')['train_acc'].transform('mean')
loss_acc_df['std_train_acc'] = loss_acc_df.groupby('epoch')['train_acc'].transform('std')
loss_acc_df['mean_test_loss'] = loss_acc_df.groupby('epoch')['test_loss'].transform('mean')
loss_acc_df['std_test_loss'] = loss_acc_df.groupby('epoch')['test_loss'].transform('std')
loss_acc_df['mean_test_acc'] = loss_acc_df.groupby('epoch')['test_acc'].transform('mean')
loss_acc_df['std_test_acc'] = loss_acc_df.groupby('epoch')['test_acc'].transform('std')
loss_acc_df = loss_acc_df[loss_acc_df.fold==1]
fig = make_subplots(rows=2, cols=1,
shared_xaxes=True,
vertical_spacing=0.05,
subplot_titles=('Loss','Accuracy'),
specs=[[{'type':'scatter'}], [{'type':'scatter'}]])
for dataset in ['train','test']:
if (dataset=='train'):
color = 'mediumblue'
elif (dataset=='test'):
color = 'crimson'
# loss (no std)
loss = go.Scatter(
x=loss_acc_df['epoch'],
y=loss_acc_df['mean_'+dataset+'_loss'],
customdata=loss_acc_df['std_'+dataset+'_loss'],
mode='markers+lines',
line={'width':2, 'color':color},
marker={'size':4, 'color':color},
name=dataset,
legendgroup=dataset,
showlegend=True,
visible=True,
hovertemplate='epoch: %{x}<br>loss: %{y:.5f}<br>sd: %{customdata:.5f}'
)
fig.add_trace(loss, row=1, col=1)
# loss (std)
upper = loss_acc_df['mean_'+dataset+'_loss'] + loss_acc_df['std_'+dataset+'_loss']
lower = loss_acc_df['mean_'+dataset+'_loss'] - loss_acc_df['std_'+dataset+'_loss']
loss = go.Scatter(
x=np.concatenate([loss_acc_df.index, loss_acc_df.index[::-1]])-loss_acc_df.index[0]+1,
y=pd.concat([upper, lower[::-1]]),
fill='toself',
mode='lines',
line={'width':0, 'color':color},
opacity=0.7,
name=dataset,
legendgroup=dataset,
showlegend=False,
visible=True,
hoverinfo='skip'
)
fig.add_trace(loss, row=1, col=1)
# acc (no std)
acc = go.Scatter(
x=loss_acc_df['epoch'],
y=loss_acc_df['mean_'+dataset+'_acc'],
customdata=loss_acc_df['std_'+dataset+'_acc'],
mode='markers+lines',
line={'width':2, 'color':color},
marker={'size':4, 'color':color},
name=dataset,
legendgroup=dataset,
showlegend=False,
visible=True,
hovertemplate='epoch: %{x}<br>acc: %{y:.5f}<br>sd: %{customdata:.5f}'
)
fig.add_trace(acc, row=2, col=1)
# acc (std)
upper = loss_acc_df['mean_'+dataset+'_acc'] + loss_acc_df['std_'+dataset+'_acc']
lower = loss_acc_df['mean_'+dataset+'_acc'] - loss_acc_df['std_'+dataset+'_acc']
acc = go.Scatter(
x=np.concatenate([loss_acc_df.index, loss_acc_df.index[::-1]])-loss_acc_df.index[0]+1,
y=pd.concat([upper, lower[::-1]]),
fill='toself',
mode='lines',
line={'width':0, 'color':color},
opacity=0.7,
name=dataset,
legendgroup=dataset,
showlegend=False,
visible=True,
hoverinfo='skip'
)
fig.add_trace(acc, row=2, col=1)
# formatting
fig.update_layout(
autosize=False,
width=800,
height=800,
margin={'l':0, 'r':0, 't':70, 'b':100},
legend={'orientation':'h',
'itemsizing':'constant',
'xanchor':'center',
'yanchor':'bottom',
'y':-0.07,
'x':0.5},
title={'text':lossacc_title,
'xanchor':'center',
'yanchor':'top',
'x':0.5,
'y':0.98},
hovermode='x')
fig['layout']['annotations'] += (
{'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.14,
'showarrow':False,
'text':lossacc_caption
},
)
plotly_config = {'displaylogo':False,
'modeBarButtonsToRemove': ['autoScale2d','toggleSpikelines','hoverClosestCartesian','hoverCompareCartesian','lasso2d','select2d']}
fig.show(config=plotly_config)
# PLOT CONFUSION MATRIX
cm = cm.astype('float32')
for i in range(len(cm)):
cm[i] = cm[i]/np.sum(cm[i]) # rows/columns total to 1
labels=clipdata_df['clip_name'].to_numpy()
plotly_data = go.Heatmap(z=np.rot90(cm.T), y=np.flip(labels), x=labels, colorscale="blues",
zmin=0, zmax=1,
hovertemplate='True: %{y}<br>Predicted: %{x}<br>p: %{z:.5f}')
annotations = []
for i, row in enumerate(cm.T):
for j, value in enumerate(row):
if (value > 0.5):
color = 'white'
else:
color = 'black'
annotations.append({
'x': labels[i],
'y': labels[j],
'font': {'color': color},
'text': str(round(value,3)),
'xref': 'x1',
'yref': 'y1',
'showarrow': False
})
annotations.append({
'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.18,
'showarrow':False,
'text':cm_caption
})
plotly_layout = go.Layout(
autosize=False,
width=800,
height=800,
margin={'l':0, 'r':0, 't':40, 'b':120},
xaxis={'title': 'Predicted', 'fixedrange':True, 'type':'category'},
yaxis={'title': 'True', 'fixedrange':True, 'type':'category'},
title={'text':cm_title,
'xanchor':'center',
'yanchor':'top',
'x':0.5,
'y':0.98},
annotations=annotations)
plotly_config = {'displaylogo':False,
'modeBarButtonsToRemove': ['autoScale2d','toggleSpikelines','hoverClosestCartesian','hoverCompareCartesian','lasso2d','select2d','zoom2d','pan2d','zoomIn2d','zoomOut2d','resetScale2d']}
fig = go.Figure(data=plotly_data, layout=plotly_layout)
fig.show(config=plotly_config)
nested_cv_lstm(
dataset=coeff_dataset,
outer_kfold=10,
num_outer_epochs=50,
inner_kfold=10,
num_inner_epochs=50,
batch_size=16,
k_hidden=[3,15,30,50,150],
k_layers=[1,2,3],
k_seed=330,
scoring='acc',
dropout=0.5,
device=torch.device('cuda:0'), #torch.device('cuda:0' if torch.cuda.is_available() else 'cpu'),
opt_title='LSTM Hyperparameter Optimization',
opt_caption='<b>Fig. 9.</b> LSTM hyperparameter optimization using 10-fold nested cross validation. (A) Mean validation accuracy.<br>(B) Relative frequency of being chosen as the best model from the inner CV.',
lossacc_title='LSTM Loss and Accuracy',
lossacc_caption='<b>Fig. 10.</b> LSTM mean loss and accuracy for train and test sets across outer folds of 10-fold nested cross validation.<br>Error bars show the standard deviation of the mean loss and accuracy at each epoch.',
cm_title='LSTM Confusion Matrix',
cm_caption='<b>Fig. 11.</b> Mean confusion matrix for trained LSTM model across outer folds of 10-fold nested cross validation.'
)
Like with the MLP, we also want to check how the model performs with shuffled labels.
nested_cv_lstm(
dataset=coeff_dataset_shuffled,
outer_kfold=10,
num_outer_epochs=50,
inner_kfold=10,
num_inner_epochs=50,
batch_size=16,
k_hidden=[30,50,150],
k_layers=[1,2,3],
k_seed=330,
scoring='acc',
dropout=0.5,
device=torch.device('cuda:0'), #torch.device('cuda:0' if torch.cuda.is_available() else 'cpu'),
opt_title='LSTM Hyperparameter Optimization (Shuffled Labels)',
opt_caption='<b>Fig. 12.</b> LSTM hyperparameter optimization using 10-fold nested cross validation with shuffled labels.<br>(A) Mean validation accuracy. (B) Relative frequency of being chosen as the best model from the inner CV.',
lossacc_title='LSTM Loss and Accuracy (Shuffled Labels)',
lossacc_caption='<b>Fig. 13.</b> LSTM mean loss and accuracy for train and test sets with shuffled labels across outer folds of<br>10-fold nested cross validation. Error bars show the standard deviation of the mean loss and accuracy at each epoch.',
cm_title='LSTM Confusion Matrix (Shuffled Labels)',
cm_caption='<b>Fig. 14.</b> Mean confusion matrix for trained LSTM model across outer folds of 10-fold nested cross validation with shuffled labels.'
)
Once again we see the model guessing “testretest” for all examples. This indicates that the original LSTM model is working as intended.
Temporal Convolutional Network (TCN)¶
LSTMs use a dynamically-changing contextual window due to it’s hidden state. We can also try convolutional networks that use a static fixed-sized window or “kernel.”
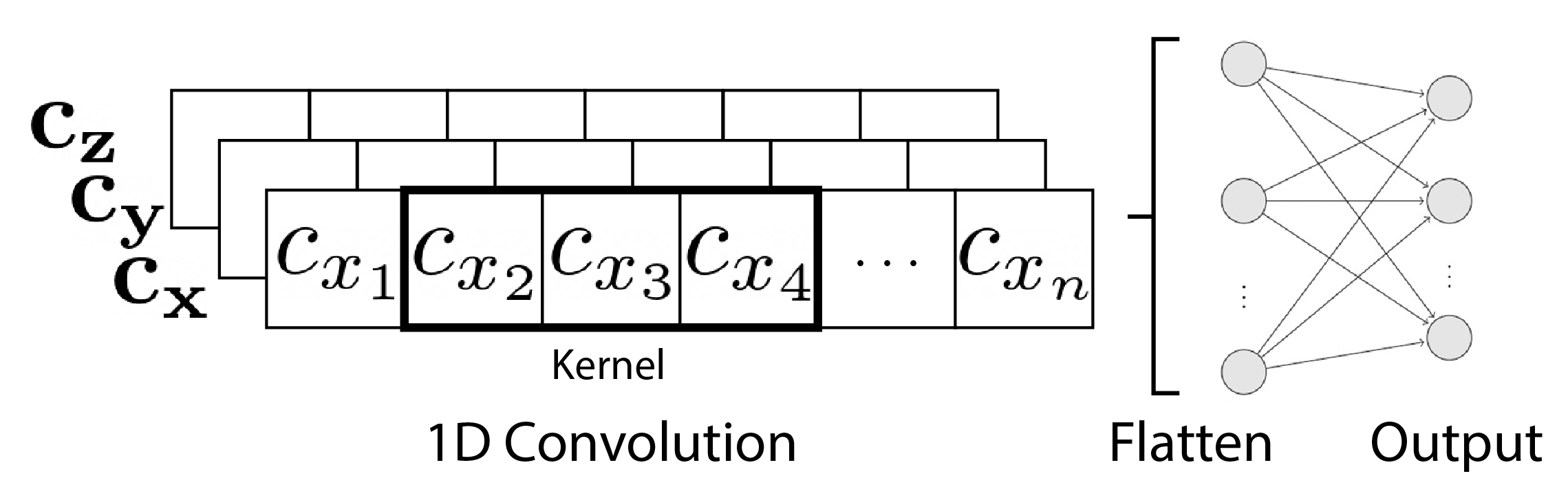
The window starts at the first basis, then moves until it reaches the end of the sequence at the last basis. There are 3 1D input sequences for each trajectory representing the x, y, and z coefficients, so the TCN will have 3 input channels. The kernel moves across each channel separately. Since the bases are ordered by time, the kernel is moving along the temporal dimension, hence the name TCN.
class TCN(nn.Module):
def __init__(self, k_input, k_hidden, k_window, k_output, dropout=0):
nn.Module.__init__(self) #super().__init__()
self.conv = nn.Conv1d(in_channels=k_input, out_channels=k_hidden, kernel_size=k_window, stride=1)
self.pad = nn.ConstantPad1d((k_window-1, 0, 0, 0), 0) # left pad zeros
self.dropout = nn.Dropout(dropout)
self.fc = nn.Linear(k_hidden*50, k_output)
def forward(self, x):
batch_size = x.shape[0]
x = x.view(batch_size,3,50) # (batch_size, in_channels, data_dim)
x = F.relu(self.conv(self.pad(x)))
x = x.flatten(1)
x = self.dropout(x)
y = self.fc(x)
return y
def grid_search_tcn(train_dl, val_dl, num_epochs, k_hidden, k_window, scoring, criterion, device, dropout=0):
val_scores = np.zeros((len(k_hidden),len(k_window)))
for i,hidden in enumerate(k_hidden):
for j,window in enumerate(k_window):
model = TCN(3,hidden,window,15,dropout)
optimizer = optim.Adam(model.parameters())
avg_val_score = 0
for epoch in range(num_epochs-5):
train(model, criterion, optimizer, train_dl, device)
for epoch in range(5):
train(model, criterion, optimizer, train_dl, device)
val_loss, val_acc = evaluate(model, criterion, val_dl, device)
# if (scoring=='acc' or scoring=='accuracy'):
# avg_val_score += val_acc
# elif (scoring=='loss'):
# avg_val_score += val_loss
avg_val_score += val_acc # no if statement for speed
avg_val_score /= 5
val_scores[i][j] = avg_val_score
return val_scores
def nested_cv_tcn(dataset, outer_kfold, num_outer_epochs, inner_kfold, num_inner_epochs, batch_size, k_hidden, k_window, k_seed, scoring, device, opt_title, opt_caption, lossacc_title, lossacc_caption, cm_title, cm_caption, dropout=0.5):
# set seed for reproducibility
torch.manual_seed(k_seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
# NESTED CV
loss_acc_df = pd.DataFrame(columns=['fold','epoch','train_loss','train_acc','test_loss','test_acc'])
x, y = dataset[:]
cm = np.zeros((torch.max(y).item()+1,torch.max(y).item()+1), dtype=np.int)
total_val_score = np.zeros((len(k_hidden),len(k_window)))
total_freq = np.zeros((len(k_hidden),len(k_window)), dtype=np.int)
outer_kfold = KFold(n_splits=outer_kfold, shuffle=True, random_state=0)
inner_kfold = KFold(n_splits=inner_kfold, shuffle=True, random_state=0)
criterion = nn.CrossEntropyLoss()
# Outer CV (trainval/test split)
current_outer_fold = 1
for trainval_index, test_index in outer_kfold.split(x, y):
print(f'Outer fold {current_outer_fold}/{outer_kfold.n_splits}')
x_trainval = x[trainval_index]
y_trainval = y[trainval_index]
x_test = x[test_index]
y_test = y[test_index]
trainval_data = TensorDataset(x_trainval, y_trainval)
test_data = TensorDataset(x_test, y_test)
# Inner CV (train/val split)
current_inner_fold = 1
inner_val_score = np.zeros((len(k_hidden),len(k_window)))
for train_index, val_index in inner_kfold.split(x_trainval, y_trainval):
print(f' Inner fold {current_inner_fold}/{inner_kfold.n_splits}')
train_data = TensorDataset(x_trainval[train_index], y_trainval[train_index])
val_data = TensorDataset(x_trainval[val_index], y_trainval[val_index])
train_dl = DataLoader(train_data, batch_size=batch_size, shuffle=True)
val_dl = DataLoader(val_data, batch_size=batch_size, shuffle=True)
fold_val_score = grid_search_tcn(train_dl, val_dl, num_inner_epochs, k_hidden, k_window, scoring, criterion, device, dropout)
inner_val_score = np.add(inner_val_score, fold_val_score)
current_inner_fold += 1
if (scoring=='acc' or scoring=='accuracy'):
best_params = np.unravel_index(np.argmax(inner_val_score, axis=None), inner_val_score.shape)
elif (scoring=='loss'):
best_params = np.unravel_index(np.argmin(inner_val_score, axis=None), inner_val_score.shape)
total_freq[best_params[0],best_params[1]] += 1
model = TCN(3,k_hidden[best_params[0]],k_window[best_params[1]],15,dropout)
optimizer = optim.Adam(model.parameters())
trainval_dl = DataLoader(trainval_data, batch_size=batch_size, shuffle=True)
test_dl = DataLoader(test_data, batch_size=batch_size, shuffle=True)
for epoch in range(1,num_outer_epochs+1):
trainval_loss, trainval_acc = train(model, criterion, optimizer, trainval_dl, device)
if (epoch==num_outer_epochs):
test_loss, test_acc, fold_cm = evaluate_cm(model, criterion, test_dl, device)
cm += fold_cm
else:
test_loss, test_acc = evaluate(model, criterion, test_dl, device)
loss_acc_df = loss_acc_df.append({
'fold':current_outer_fold,
'epoch':epoch,
'train_loss':trainval_loss.item(),
'train_acc':trainval_acc.item(),
'test_loss':test_loss.item(),
'test_acc':test_acc.item()},
ignore_index=True)
current_outer_fold += 1
total_val_score = np.add(total_val_score, inner_val_score)
# PLOT HYPERPARAMETERS
clear_output()
avg_val_score = total_val_score / (outer_kfold.n_splits * inner_kfold.n_splits)
avg_freq = total_freq / outer_kfold.n_splits
plotly_data = []
plotly_data.append(
go.Heatmap(z=np.flip(avg_val_score,axis=0), y=np.flip(np.array(k_hidden)), x=k_window, colorscale='blues',
zmin=np.amin(avg_val_score), zmax=np.amax(avg_val_score),
name='Validation<br>Accuracy',
customdata=np.flip(avg_freq,axis=0),
hovertemplate='k_hidden: %{y}<br>k_window: %{x}<br>'+scoring+': %{z:.5f}<br>rel freq: %{customdata:.3f}',
visible=True))
plotly_data.append(
go.Heatmap(z=np.flip(avg_freq,axis=0), y=np.flip(np.array(k_hidden)), x=k_window, colorscale='blues',
zmin=np.amin(avg_freq), zmax=np.amax(avg_freq),
name='Relative<br>Frequency',
customdata=np.flip(avg_val_score,axis=0),
hovertemplate='k_hidden: %{y}<br>k_window %{x}<br>'+scoring+': %{customdata:.5f}<br>rel freq: %{z:.3f}',
visible=False))
acc_annotations = []
mid = (np.amin(avg_val_score)+np.amax(avg_val_score))/2
for i, row in enumerate(avg_val_score.T):
for j, value in enumerate(row):
if (value > mid):
color = 'white'
else:
color = 'black'
acc_annotations.append({
'x': k_window[i],
'y': k_hidden[j],
'font': {'color': color},
'text': str(round(value,5)),
'xref': 'x1',
'yref': 'y1',
'showarrow': False
})
acc_annotations.append({
'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.14,
'showarrow':False,
'text':opt_caption
})
freq_annotations = []
mid = (np.amin(avg_freq)+np.amax(avg_freq))/2
for i, row in enumerate(avg_freq.T):
for j, value in enumerate(row):
if (value > mid):
color = 'white'
else:
color = 'black'
freq_annotations.append({
'x': k_window[i],
'y': k_hidden[j],
'font': {'color': color},
'text': str(round(value,3)),
'xref': 'x1',
'yref': 'y1',
'showarrow': False
})
freq_annotations.append({
'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.14,
'showarrow':False,
'text':opt_caption
})
plotly_layout = go.Layout(
autosize=False,
width=800,
height=800,
margin={'l':0, 'r':0, 't':40, 'b':100},
xaxis={'title': 'k_window', 'fixedrange':True, 'type':'category'},
yaxis={'title': 'k_hidden', 'fixedrange':True, 'type':'category'},
title={'text':opt_title,
'xanchor':'center',
'yanchor':'top',
'x':0.5,
'y':0.98},
annotations=acc_annotations,
updatemenus=[{'type':'buttons',
'direction':'left',
'pad':{'l':0, 'r':0, 't':0, 'b':0},
'xanchor':'left',
'yanchor':'top',
'x':0,
'y':1.055,
'buttons':[
{'label':'Val. Acc.',
'method': 'update',
'args':[{'visible': [True,False]},
{'annotations': acc_annotations}]},
{'label':'Rel. Freq.',
'method': 'update',
'args':[{'visible': [False,True]},
{'annotations': freq_annotations}]}
]}])
plotly_config = {'displaylogo':False,
'modeBarButtonsToRemove': ['autoScale2d','toggleSpikelines','hoverClosestCartesian','hoverCompareCartesian','lasso2d','select2d','zoom2d','pan2d','zoomIn2d','zoomOut2d','resetScale2d']}
fig = go.Figure(data=plotly_data, layout=plotly_layout)
fig.show(config=plotly_config)
# PLOT LOSS / ACCURACY
loss_acc_df['mean_train_loss'] = loss_acc_df.groupby('epoch')['train_loss'].transform('mean')
loss_acc_df['std_train_loss'] = loss_acc_df.groupby('epoch')['train_loss'].transform('std')
loss_acc_df['mean_train_acc'] = loss_acc_df.groupby('epoch')['train_acc'].transform('mean')
loss_acc_df['std_train_acc'] = loss_acc_df.groupby('epoch')['train_acc'].transform('std')
loss_acc_df['mean_test_loss'] = loss_acc_df.groupby('epoch')['test_loss'].transform('mean')
loss_acc_df['std_test_loss'] = loss_acc_df.groupby('epoch')['test_loss'].transform('std')
loss_acc_df['mean_test_acc'] = loss_acc_df.groupby('epoch')['test_acc'].transform('mean')
loss_acc_df['std_test_acc'] = loss_acc_df.groupby('epoch')['test_acc'].transform('std')
loss_acc_df = loss_acc_df[loss_acc_df.fold==1]
fig = make_subplots(rows=2, cols=1,
shared_xaxes=True,
vertical_spacing=0.05,
subplot_titles=('Loss','Accuracy'),
specs=[[{'type':'scatter'}], [{'type':'scatter'}]])
for dataset in ['train','test']:
if (dataset=='train'):
color = 'mediumblue'
elif (dataset=='test'):
color = 'crimson'
# loss (no std)
loss = go.Scatter(
x=loss_acc_df['epoch'],
y=loss_acc_df['mean_'+dataset+'_loss'],
customdata=loss_acc_df['std_'+dataset+'_loss'],
mode='markers+lines',
line={'width':2, 'color':color},
marker={'size':4, 'color':color},
name=dataset,
legendgroup=dataset,
showlegend=True,
visible=True,
hovertemplate='epoch: %{x}<br>loss: %{y:.5f}<br>sd: %{customdata:.5f}'
)
fig.add_trace(loss, row=1, col=1)
# loss (std)
upper = loss_acc_df['mean_'+dataset+'_loss'] + loss_acc_df['std_'+dataset+'_loss']
lower = loss_acc_df['mean_'+dataset+'_loss'] - loss_acc_df['std_'+dataset+'_loss']
loss = go.Scatter(
x=np.concatenate([loss_acc_df.index, loss_acc_df.index[::-1]])-loss_acc_df.index[0]+1,
y=pd.concat([upper, lower[::-1]]),
fill='toself',
mode='lines',
line={'width':0, 'color':color},
opacity=0.7,
name=dataset,
legendgroup=dataset,
showlegend=False,
visible=True,
hoverinfo='skip'
)
fig.add_trace(loss, row=1, col=1)
# acc (no std)
acc = go.Scatter(
x=loss_acc_df['epoch'],
y=loss_acc_df['mean_'+dataset+'_acc'],
customdata=loss_acc_df['std_'+dataset+'_acc'],
mode='markers+lines',
line={'width':2, 'color':color},
marker={'size':4, 'color':color},
name=dataset,
legendgroup=dataset,
showlegend=False,
visible=True,
hovertemplate='epoch: %{x}<br>acc: %{y:.5f}<br>sd: %{customdata:.5f}'
)
fig.add_trace(acc, row=2, col=1)
# acc (std)
upper = loss_acc_df['mean_'+dataset+'_acc'] + loss_acc_df['std_'+dataset+'_acc']
lower = loss_acc_df['mean_'+dataset+'_acc'] - loss_acc_df['std_'+dataset+'_acc']
acc = go.Scatter(
x=np.concatenate([loss_acc_df.index, loss_acc_df.index[::-1]])-loss_acc_df.index[0]+1,
y=pd.concat([upper, lower[::-1]]),
fill='toself',
mode='lines',
line={'width':0, 'color':color},
opacity=0.7,
name=dataset,
legendgroup=dataset,
showlegend=False,
visible=True,
hoverinfo='skip'
)
fig.add_trace(acc, row=2, col=1)
# formatting
fig.update_layout(
autosize=False,
width=800,
height=800,
margin={'l':0, 'r':0, 't':70, 'b':100},
legend={'orientation':'h',
'itemsizing':'constant',
'xanchor':'center',
'yanchor':'bottom',
'y':-0.07,
'x':0.5},
title={'text':lossacc_title,
'xanchor':'center',
'yanchor':'top',
'x':0.5,
'y':0.98},
hovermode='x')
fig['layout']['annotations'] += (
{'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.14,
'showarrow':False,
'text':lossacc_caption
},
)
plotly_config = {'displaylogo':False,
'modeBarButtonsToRemove': ['autoScale2d','toggleSpikelines','hoverClosestCartesian','hoverCompareCartesian','lasso2d','select2d']}
fig.show(config=plotly_config)
# PLOT CONFUSION MATRIX
cm = cm.astype('float32')
for i in range(len(cm)):
cm[i] = cm[i]/np.sum(cm[i]) # rows/columns total to 1
labels=clipdata_df['clip_name'].to_numpy()
plotly_data = go.Heatmap(z=np.rot90(cm.T), y=np.flip(labels), x=labels, colorscale="blues",
zmin=0, zmax=1,
hovertemplate='True: %{y}<br>Predicted: %{x}<br>p: %{z:.5f}')
annotations = []
for i, row in enumerate(cm.T):
for j, value in enumerate(row):
if (value > 0.5):
color = 'white'
else:
color = 'black'
annotations.append({
'x': labels[i],
'y': labels[j],
'font': {'color': color},
'text': str(round(value,3)),
'xref': 'x1',
'yref': 'y1',
'showarrow': False
})
annotations.append({
'xref':'paper',
'yref':'paper',
'xanchor':'center',
'yanchor':'bottom',
'x':0.5,
'y':-0.18,
'showarrow':False,
'text':cm_caption
})
plotly_layout = go.Layout(
autosize=False,
width=800,
height=800,
margin={'l':0, 'r':0, 't':40, 'b':120},
xaxis={'title': 'Predicted', 'fixedrange':True, 'type':'category'},
yaxis={'title': 'True', 'fixedrange':True, 'type':'category'},
title={'text':cm_title,
'xanchor':'center',
'yanchor':'top',
'x':0.5,
'y':0.98},
annotations=annotations)
plotly_config = {'displaylogo':False,
'modeBarButtonsToRemove': ['autoScale2d','toggleSpikelines','hoverClosestCartesian','hoverCompareCartesian','lasso2d','select2d','zoom2d','pan2d','zoomIn2d','zoomOut2d','resetScale2d']}
fig = go.Figure(data=plotly_data, layout=plotly_layout)
fig.show(config=plotly_config)
nested_cv_tcn(
dataset=coeff_dataset,
outer_kfold=10,
num_outer_epochs=50,
inner_kfold=10,
num_inner_epochs=50,
batch_size=16,
k_hidden=[3,15,30,50,100,150],
k_window=[3,5,10,20,30],
k_seed=330,
scoring='acc',
dropout=0.5,
device=torch.device('cpu'), #torch.device('cuda:0' if torch.cuda.is_available() else 'cpu'),
opt_title='TCN Hyperparameter Optimization',
opt_caption='<b>Fig. 15.</b> TCN hyperparameter optimization using 10-fold nested cross validation. (A) Mean validation accuracy.<br>(B) Relative frequency of being chosen as the best model from the inner CV.',
lossacc_title='TCN Loss and Accuracy',
lossacc_caption='<b>Fig. 16.</b> TCN mean loss and accuracy for train and test sets across outer folds of 10-fold nested cross validation.<br>Error bars show the standard deviation of the mean loss and accuracy at each epoch.',
cm_title='TCN Confusion Matrix',
cm_caption='<b>Fig. 17.</b> Mean confusion matrix for trained TCN model across outer folds of 10-fold nested cross validation.'
)
nested_cv_tcn(
dataset=coeff_dataset_shuffled,
outer_kfold=10,
num_outer_epochs=50,
inner_kfold=10,
num_inner_epochs=50,
batch_size=16,
k_hidden=[100,150],
k_window=[3,5,10],
k_seed=330,
scoring='acc',
dropout=0.5,
device=torch.device('cpu'), #torch.device('cuda:0' if torch.cuda.is_available() else 'cpu'),
opt_title='TCN Hyperparameter Optimization (Shuffled Labels)',
opt_caption='<b>Fig. 18.</b> TCN hyperparameter optimization using 10-fold nested cross validation with shuffled labels.<br>(A) Mean validation accuracy. (B) Relative frequency of being chosen as the best model from the inner CV.',
lossacc_title='TCN Loss and Accuracy (Shuffled Labels)',
lossacc_caption='<b>Fig. 19.</b> TCN mean loss and accuracy for train and test sets with shuffled labels across outer folds of<br>10-fold nested cross validation. Error bars show the standard deviation of the mean loss and accuracy at each epoch.',
cm_title='TCN Confusion Matrix (Shuffled Labels)',
cm_caption='<b>Fig. 20.</b> Mean confusion matrix for trained TCN model across outer folds of 10-fold nested cross validation with shuffled labels.'
)
